Bio-Loom & QSAR Web Database
Introduction
Merck's voluntary withdrawal of Vioxx (rofecoxib) came as a surprise to many observers of the pharmaceutical scene. It serves notice, if one were needed, that even careful observation of clinical results may not disclose a relatively rare but devastating side-effect that becomes evident only after millions of prescriptions have been written. In recent weeks there has undoubtedly been an intense review of the many steps taken in the development of Vioxx and similar cox-2 inhibitors (e.g. Celebrex, celecoxib, Bextra, valdecoxib) to see if there were any hints that were overlooked that coronary problems would necessitate its removal from the anti-inflammatory market. Even the voluminous research records in Merck's labs may not provide such a hint, but it still can be useful and informative for medicinal chemists to review the published literature to trace cox-2 inhibitor development and speculate on how many and what kinds of 'decision points' were encountered along the way. In a featured article entitled "Tough Road for Agrochemicals" (Chemical and Engineering News, 9/20/04, p.48) John Lyga of FMC is quoted: "Valuable scientific lessons can be derived from open discussion of projects that never made it to the marketplace." Certainly this statement holds true for pharmaceuticals as well. This summarized tutorial is intended to illustrate how quickly and effectively users of BioByte's new Bio-Loom and QSAR products can access the pertinent data for such discussions. In the references found in this search, the authors present some of rationale for the pathways taken in following early leads, but, as we shall see, QSAR's meaningful physico-chemical parameters often provide additional perspectives of the mechanisms of the bioactivity involved.
|
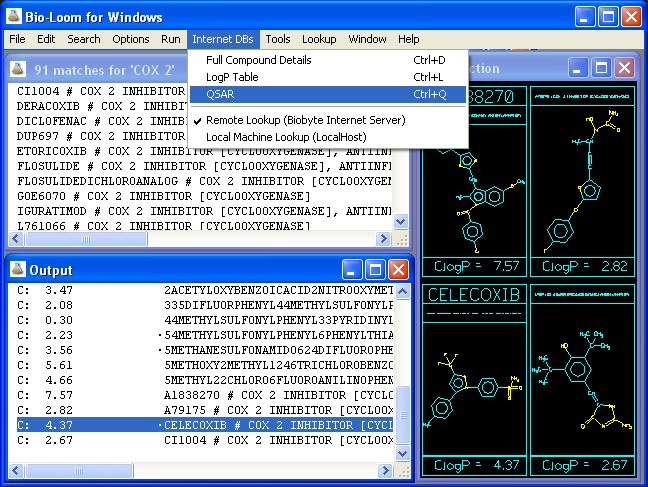
Figure 1: QSAR database lookup from Bio-Loom
Pre-Scan Using Bio-Loom
Note: Portions of the following discussion will be accessible only to users of the Bio-Loom program and are not depicted herein; these will appear in maroon.
A user would begin this Tutorial with the Bio-Loom 'Lookup' menu set at 'Activity', the 'Options' set at 'terse', and cox-2 typed in the Input Panel. After selecting the 'Run' command (Ctl+R), a list of 114 hits appears in the 'Matches' Window, sorted on a highlighted cox-2. The first two dozen are mixed cox-1 & 2 inhibitors with other activities (e.g. leukotriene, lipoxygenase, & PAF action) listed later on each line. Sort by name is also available as an option in the Tools menu. The structures of the first dozen or so should be scanned (four or six at a time, as seen in Figure 1) using the 'Run Page' command (Ctrl+G). Besides those having names with the familiar suffix, '-coxib', there are many preceded by a 'bullet' indicating they are present in the Hansch database of QSAR equations. Later, when these are accessed in the QSAR Search program, one will find the measured activity for each (usually as IC-50) together with the physico-chemical descriptors that 'explain' the activity difference in that set
Even from a quick scan of structures, it is apparent that the basic 'lead' in 'coxib' development is a central ring (often five-membered and unsaturated, or heteroaromatic) with two aromatic rings attached to it. One of these aromatic rings consists of a p-phenylsulfone and the other is often a phenyl substituted with a halogen or methyl. More detailed discussions of this 'lead scaffold' will appear in the references cited in the results of the QSAR Search which will be performed next, and almost all 'lead scaffolds' are present in the 114 'hit file'. Of course, if a more detailed study is contemplated, these journal references can be accessed to find interesting observations, such as the Merck group's finding that halogen substitution of the 3-phenyl ring increases the selectivity of cox-2 over cox-1, and that of Shiu et.al. that celecoxib (Celebrex) with a pyrrazole central ring is largely metabolized by the P-450 suite of enzymes, while the furanone ring of refecoxib (Vioxx) is metabolized by unidentified (in the year 2000) cytosolic enzymes.1
|
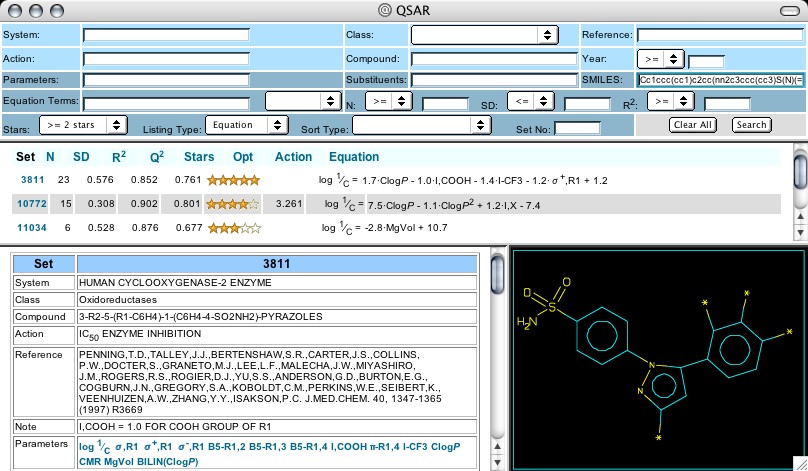
Figure 2: QSAR Web Search
QSAR Equations
The initial survey step using Bio-Loom can be concluded by selecting the name celecoxib in the Output window, (which has a bullet), clicking on 'internet DBs' in the upper menu bar, and selecting 'QSAR' (Figure 1). After the net is accessed (takes a few seconds) a Search panel is displayed with the SMILES of celecoxib (Celebrex) in the proper window. Three Sets (with their equations) are shown in the middle panel: 3811, 10772, & 11034 (Figure 2).
|
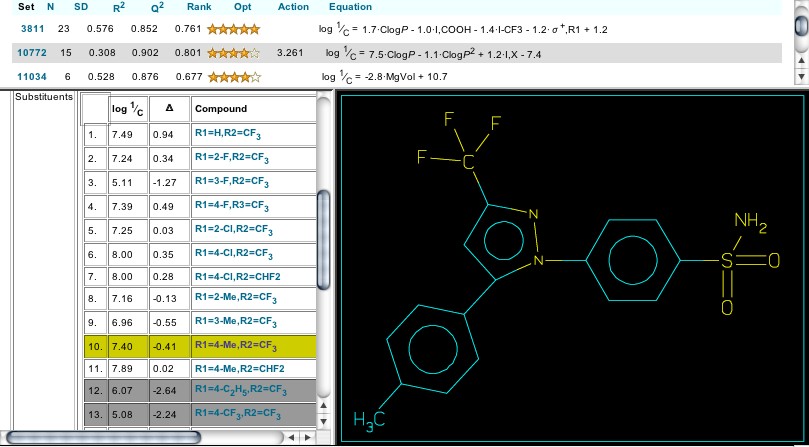
Figure 3: Celebrex
|
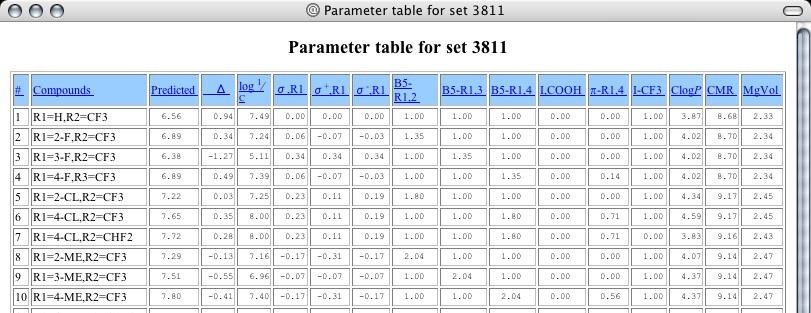
Clicking on the number 3811 delivers a lower panel (Figure 2) showing the parent structure for this set and the positions that are substituted in the analogs studied as cox-2 inhibitors by T. D. Penning et al. Cpd. #10, highlighted in yellow, is Celebrex, and its entire structure is shown when the cursor is placed over its substituent listing (Figure 3). The cox-2 inhibiting action (7.40) is expressed as log 1/C (so higher numbers show higher activity) and this appears in the first column to the right of the Cpd #. It is apparent that several analogs show higher activity than Celebrex in this simple enzyme test. Scrolling down, one notes that grey blocks denote 'outliers' to the selected equation with the deviation shown as the amount the calculation differs from the observed. Clicking on any substituent delivers a Parameter Table that shows the numerical value of each parameter used as well as the predicted activity, as seen below:
|
|
As this Tutorial continues, it will become evident that hydrophobicity (CLOGP) plays an important role in cox-2 inhibition, sometimes in parabolic or bilinear fashion. This is unusual in isolated enzyme activity. Also, the significance of the sigma-plus parameter in the equation for Set 3811 deserves further comment. Although sigma-plus was designed to apply to reactions where resonance delocalization of a positive charge plays a strong role, nevertheless it is strongly correlated with radical reaction mechanisms.2 There are 11 equations in the database that delineate the significance of σ+ in the interaction of molecules with the cox-2 receptor. The Penning reference also reports the data for cox-1 inhibition (analyzed in Set 3813), and thus some factors contributing to selectivity of cox-2 over cox-1 can be assessed.3
|
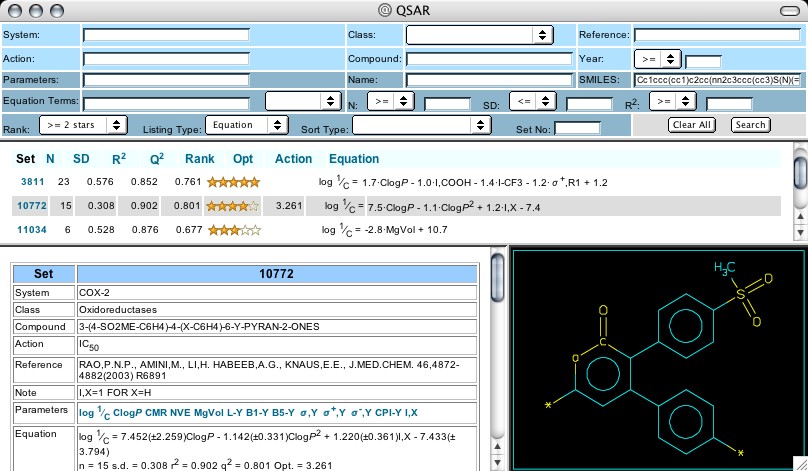
Figure 4: QSAR set 10772
|
Clicking on the Set No. 10772, one notes that it investigates the cox-2 activities of analogs of a parent where the central ring is a pyran-2-one, substituted at the 3-position with (p-methylsulfonyl)phenyl (Figure 4). The coefficient for the indicator variable, I-X (+1.22) tells us that when the phenyl at the 4-position is p- substituted, activity is decreased. The activities for both Celebrex (7.24) and Vioxx (6.37) are listed for comparison with the pyranone analogs, but neither is included in the equation (Figure 5). An optimal hydrophobicity (log P = 3.26) is seen for the variations in this set. Note that Set 11034 contains celecoxib and valdecoxib (Bextra), but it deals with their activity as carbonic anydrase inhibitors, comparing them with acetazolamide etc. This may be important information in assessing its side-reactions, wanted or unwanted.
|
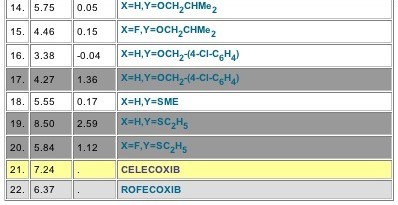
Figure 5: Celebrex (CELECOXIB) and Vioxx (ROFECOXIB) cox-2 activities in set 10772
|
To continue the Tutorial, clear the Input by clicking on the 'Reset' button (directly beneath the Input Window), and enter cox-2 in the 'System' panel. The first of the 58 hits (Set 1042) studies a variety of older NSAID structures that display mixed cox-1 and cox-2 inhibitory activity. DUP-697 in this set and flurbuprofen and tolfunamic acid in Set 3639 show cox-2 inhibition activity levels on a par with many of the newer 'coxib' drugs. Hydrophobicity and pKa are the important parameters noted.
|
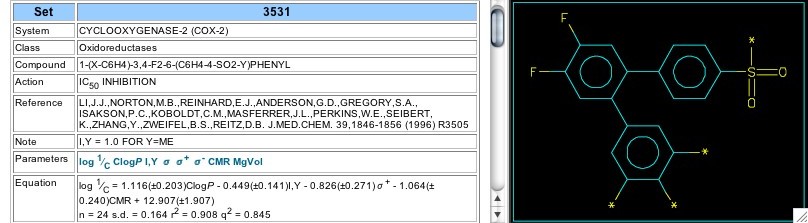
Figure 6: QSAR set 3531
|
Some of the Searle-(Pfizer) group's early work (1996) leading to Celebrex is shown in Set 3531, where the central ring is a difluorophenyl (Figure 6). The coefficient of the indicator variable (-0.45) shows that the sulfonamido group (the moiety ending up on Celebrex) is slightly more active than methylsulfonyl. The opposing coefficients for CLOGP and CMR show a kind of parabolic dependence upon the size/hydrophobicity combination of the substituents on the other 'outer' phenyl ring.
|
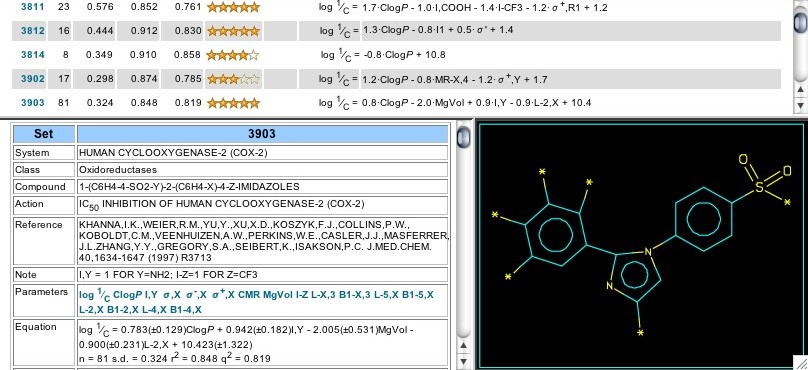
Figure 7: Sets 3811-3903
|
In later reports by this group (1997; Sets 3811, 3814) the parent has a pyrazole as the central ring; for Set 3902 it is pyrrole, and for Set 3903 it is imidazole (Figure 7). In Set 3811 (mentioned earlier) the 3-CHF2 substituent on the pyrazole appears to add more activity than the 3-CF3, but the latter was retained in Celebrex. (Perhaps to avoid complications of the activated hydrogen?) Set 3814 shows that the sulfone moiety appears to perform about as well on the phenyl attached to the N-1 nitrogen of the pyrazole as it does if placed on a phenyl at C-5. Indeed, the sulfone group can be replaced with methoxyl or with chlorine without much change in activity (Cpds. 6, 7, & 8). Set 3903 again shows a preference for sulfonamido over methylsulfonyl (+0.94 I,Y), as does Set 3906 (+0.36 I,Y) where the central ring is an unsaturated, strained carbocycle. On the other phenyl ring the use of Verloop's steric parameters shows that there is a dependence on the width of the para substituent (B1-4) and the length of the metas (L-3).
|
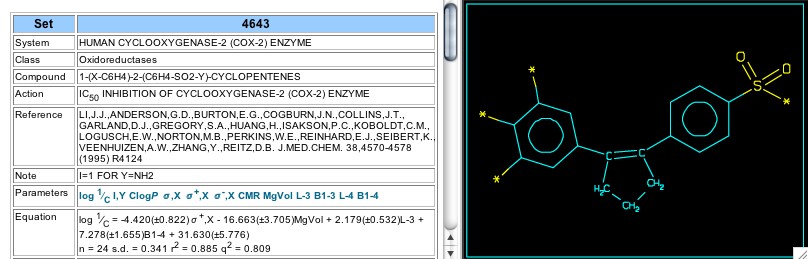
Figure 8: Sets 4643
|
In Set 4643 (Figure 8) the 'outer' phenyl rings are attached to the unsaturated carbons of cyclopentene, and for this set there was no apparent difference between methylsulfonyl and sulfonamido. As in Set 3811, there is a significant negative coefficient for the sigma-plus parameter--a hint that inhibition of cox-2 may be influenced by a resonance-suppported positive charge or radical.
|
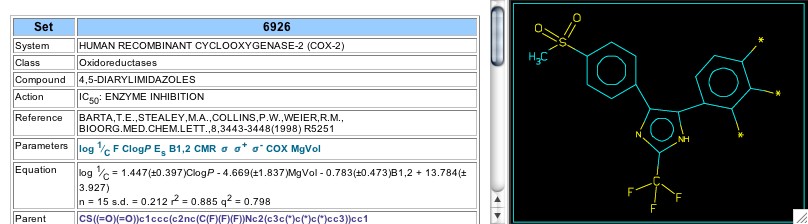
Figure 9: Set 6926
|
Further work by the Searle-(Pfizer) group (1998) using an imidazole central ring is analyzed in Set 6926 (Figure 9). The coefficients for CLOGP and volume, which are positive and negative respectively, indicate an optimal size for these analogs. Analysis of later work, Set 7140 (2000), shows a similar optimum, with the CMR parameter indicating a limiting size. In Set 6712 a series with oxazole as the central ring was studied, showing a dependence on overall hydrophobicity and sigma-plus for substituents on the 5-phenyl ring. Analysis of a smaller set (6713) confirms the sigma-plus dependence, while Set 6714, which has variation limited to the 2-position of oxazole, shows dependence on CLOGP and steric width of the 2-substituent.
|
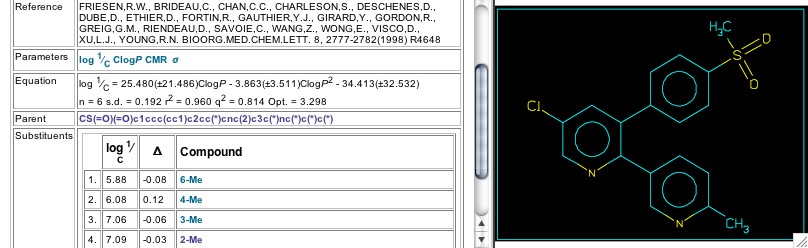
Figure 10: Arcoxia in set 5629
|
In the QSAR search illustrated by this Tutorial, the earliest published report by the Merck group appears in Set 5629 (Figure 10) where pyridine serves as the central heterocycle. It is interesting to note that Arcoxia, the cox-2 inhibitor now in clinical trials and the one Merck has announced as the (hopeful) successor to Vioxx, is Cpd. 4 in this set. Of course with substituent variation limited to just three different alkyl groups and difference in position, the resultant equation yields little information, except a hint that the optimal log P is a little over three as seen in Set 10772 above. Further work by the Merck group, using pyridine as the central ring, is seen in Sets 8385, & 8386, where Arcoxia is Cpd. 11 in both sets. Question: Does a less reactive central ring give a greater margin of safety?
|
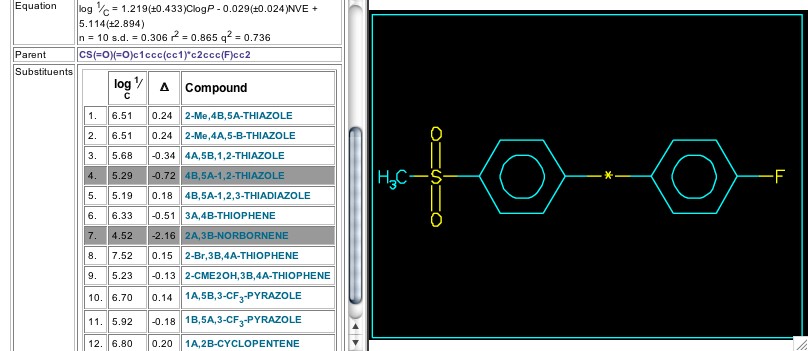
Figure 11: Substituents in set 6869
|
Set 6869 contains the reports from the Merck group on cox-2 inhibitory activity of candidates containing a wide variety of central rings (Figure 11). In Cpds. 1-4 they are thiazoles; in Cpd. 5, thiadiazole; in Cpds. 6, 8, & 9 they are thiophenes; in Cpds. 10 & 11, pyrazoles; and Cpds 7 & 12 are cycloalkenes. In view of all this structural variety, it is surprising that CLOGP by itself yields such a good correlation, especially since the experiment was done using recombinant human cox-2, and passage through cell membranes is not a factor. In more recent reports (2003) Sets 10243 and 10244 show the pyridine of the Arcoxia analogs replaced by pyridazinone.
|
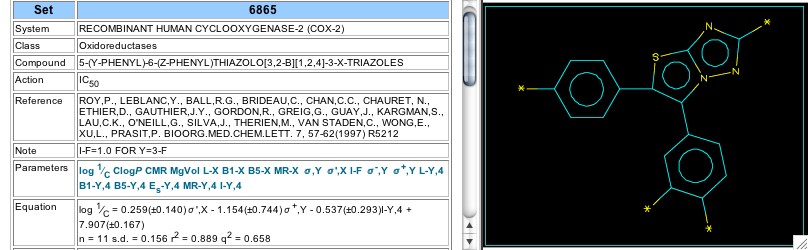
Figure 12: Set 6865
|
About the same time (1997) that the Merck group were investigating the pyridine-type cox-2 inhibitors they also reported on results using two types of thiazolo-fused rings. Set 6865 (Figure 12) analyzes those with a thiazolo-triazole central ring, and Set 6866 has an imidazothiazole fuzed ring. In neither set was a role for hydrophobicity evident, but the equation for Set 6865 hints at two electronic effects: resonance stabilization of a positive charge and an inductive effect.
|
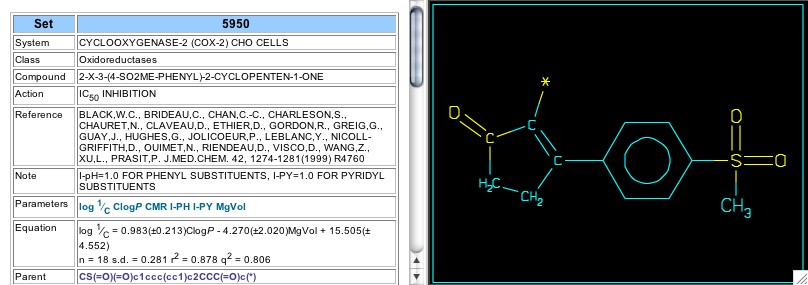
Figure 13: Equation for set 5950
|
Set 5950 includes data from a Merck publication in 1999 and features a penten-2-one central ring, which is very similar to the lactone chosen for Vioxx (Figure 13). Surprisingly, a satisfactory equation does not seem to need an electronic term for the aromatic ring attached next to the pentenone carbonyl, and Cpd. 18 fits even though the phenoxy oxygen breaks the conjugation with the central ring (Figure 14). About this same time the Merck group reported work with the central ring consisting of a benzopentanone or isobenzofuran-1-one (Sets 6842, 6843, & 6851), but these are beyond the scope of this Tutorial.
|
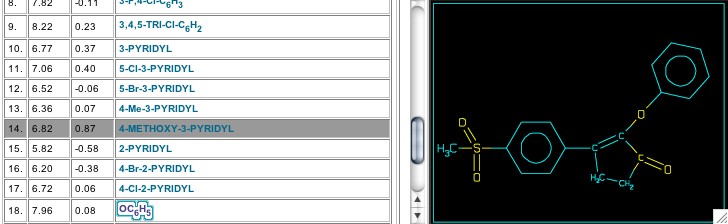
Figure 14: Compound 18 in set 5950
|
Sets 10441 and 10442 show recent Merck reports (2003) on drug candidates with the same central furan-2-one central ring as in Vioxx, but substituted at the 5-position with methyl and either hydroxyl or alkoxyl. In Set 10441 the coefficient of the indicator variable (-0.45) shows the hydroxyl to be less active, but this is less significant in Set 10442 where a sigma-plus term is justified (Figure 15).
|
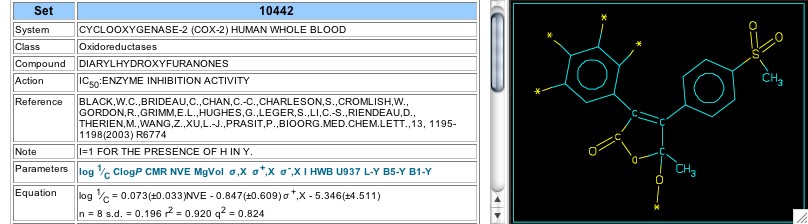
Figure 15: Set 10442
|
Obviously, the equations and the data uncovered thus far in this Tutorial QSAR Search could be examined in much greater detail, but one should not expect answers to 'pop out'. Instead one would be happy if such a survey would raise some worthwhile questions. The fact that in the most promising candidates the sulfone moiety is conjugated with a reactive central ring, and that an electronic effect (+ charge or radical stabilization) appears important poses the question: Is this contributing to cox-2 inhibition, or to the cardiotoxicity side effect, or to both?
|
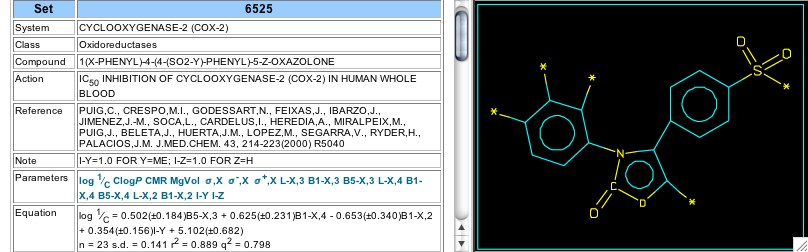
Figure 16: Set 6525
|
Some other research teams studied different reactive central rings other than those discussed above. In 2000 Puig et al reported results for over two dozen cox-2 inhibitors having an oxazol-2-one central ring, and these are seen in Set 6525 (Figure 16). The variation in this set did not disclose any hydrophobic dependence, or any electronic effect of substituents on the phenyl attached to the oxazolone nitrogen. The steric requirements of these substituents, evidenced by Verloop's width parameters, gives a hint that there is steric reaction at the reactive site. For these congeners, the methyl sulfones are more active than the sulfonamides. Any activity effect that methyl substitution of the oxazolone ring had on activity was not discernable.
|
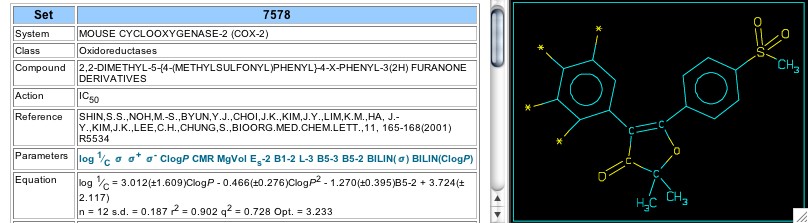
Figure 17: Set 7578
|
In 2001 Shiu et al reported on cox-2 inhibitors using 2-H-furanones as the central ring (Set 7578). This set shows a parabolic dependence on hydrophobicity, with an optimal CLOGP of 3.23 (Figure 17). In later work on similar analogs (2004; Set 11309) an optimal log P was not supported. Set 11311 analyzes the activity of spiro derivatives of these furanones, and the electronic parameter, NVE, gives a good correlation by itself (Figure 18). Set 11312 shows that replacement of the second phenyl (attached between the carbonyl and the double bond of the furanone) with a heterocycle decreases activity. In Set 11313 both phenyl rings are substituted with either chlorine or fluorine, with the unexpected result that molar volume (calc. via McGowan's method) correlates 7 of the 8 analogs very well without the need of an electronic parameter.
|
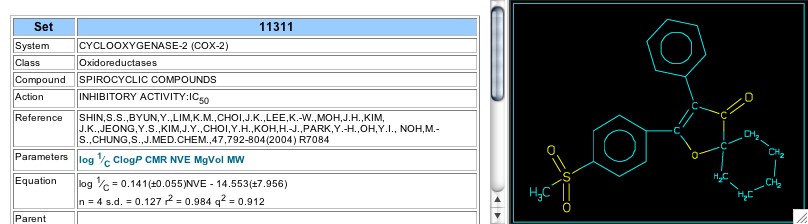
Figure 18: Set 11311
Conclusions
During the development of a drug, such as the cox-2 inhibitor, Vioxx, decisions must be made at hundreds of 'forks in the road'. Even a slight increase of the odds of making the correct choice at each step would increase the odds of final success. A few groups have told us that the use of Hansch-Fujita QSAR methodology has, indeed, increased (or sped up) the successful process of lead optimization. The retrospective view of cox-2 inhibitor development, presented in this Tutorial, illustrates a few of the critical steps where every analytical tool's information should be welcomed. When one considers that the Hansch Bio QSAR database contains over 5700 equations dealing with enzymes in general and 2600 dealing with specific receptors, one realizes that this cox-2 study is only one example of many thousands that could have been profitably examined.
The term Data Mining has come into widespread use in the last few years and is an appropriate designation for the type of exercise described in this Tutorial. The first successful diamond mining operations depended heavily on the trained human eye to detect the raw gems amid the crushed and washed Kimberlite ore. The 'rejects' were piled up as 'tailings'. After it was discovered that X-ray fluorescence made the raw gems much easier to detect, the tailings became a very valuable property and were carefully reworked. How many pharmaceutical or pesticide 'gems' lie hidden in lab notebooks waiting to be reworked? Can C-QSAR 'light up' some old, 'discarded' data the way X-rays light up 'discarded' diamonds in Kimberlite tailings?
|
-
(a) "Exploring QSAR" Hansch & Leo; ACS Reference Books, 1995; p.16.
(b) C. D. Selassie, et al., Chem Rev., 2002, 1585-2606.
|
|
